TL;DR: Guidelines in a Nutshell
General Information
- Important lab information (server access, responsibilities,…) can be found on our cloud in the folder
Lab_Orga/Important_Information. - When sharing documents, please use use the following syntax for the file name:
lastname_topic_YYYYMMDD.docx. Do not usev1,latestor similar suffixes. - An overview of geo-spatial data sets is available in our geodata wiki.
- To fulfill the requirement of a retention period of 10 years for all projects, follow our rules how to set up, maintain, and archive projects (see below).
Projects
- Create a new project for every study (i.e. publication).
- Name project folder using our unified syntax:
species_where_what_who.
→ e.g.sciurus_vulgaris_de_camtrap_kramer_s - Comply with our obligatory project folder structure.
→ easily to set up with our d6 R package:d6::new_project(). - Track and control changes of source codes: set up version control.
- For a corporate chart style, please use our d6 ggplot2 theme:
d6::theme_d6(). - Transfer your repository to our GitHub organization at the end of a project.
- Ask Moritz Wenzler-Meya (wenzler[at]izw-berlin.de) for help if needed.
Advice for Students (BSc | MSc | PhD)
- Plan regular supervisory meetings at least 3 months ahead.
- Prepare for the meeting! Collect recent achievements, potential problems, and open questions. Provide a timeline for upcoming tasks.
- Write the minutes of the meeting and send it to your supervisor(s).
- Start writing your thesis early enough and attend our “write now” meetings.
- Regularly send drafts to your supervisor(s) to get feedback.
- Allow for sufficient time to read and respond (2-4 weeks).
Table of Content
- General Information
- Conducting BSc/MSc/PhD Projects
- Organizing Workflows
- Project Folders
- Data Backup
- Manuscript Submissions/Revisions
- GitHub repository
- Appendices

General Information
The department is composed of three teams on (1) Individual Dynamics, (2) Population Dynamics and (3) Biodiversity Dynamics.
Stephanie Kramer-Schadt is the department head and leader of the Population Dynamics team. Susanne Braun is the assistant of the department head; she can assist with administrative duties (e.g. contracts, travel sheets, etc.). Moritz Wenzler-Meya and Jan Axtner are responsible for data management, data storage and the working group’s R code collection. Dagmar Thierer and Stephan Karl are the lab technicians. As each department member has a specific area of expertise, we created an expert list (see 1.3) if you need help on specific topics. PhD students and scientists are obliged to update this expert list with their own skills and responsibilities as well.
For information on the Department, check our webpage or subscribe to our Bluesky account (@ecodynizw.bsky.social) for news about papers, helpful R-code, courses or scientific positions.
Teams of the Department of Ecological Dynamics:
- Individual Dynamics – PI: Sarah Benhaiem & Sonja Metzger (field coordination)
- Population Dynamics – PI: Stephanie Kramer-Schadt & Viktoriia Radchuk
- Biodiversity Dynamics – PI: Andreas Wilting & Rahel Sollmann
Administrative support
Susanne Braun
Tel: -471
Email: assist6[at]izw-berlin.de or s.braun@izw-berlin.de
Data Managers
Dr. Jan Axtner
Tel: -342
Email: axtner[at]izw-berlin.de
Moritz Wenzler-Meya
Tel: -344
Email: wenzler[at]izw-berlin.de
In-house experts / scientists / responsibilities
Who does what in the department? Tasks and expertise as well as meeting dates and protocols can be found in the IZW Wolke.
Conducting BSc/MSc/PhD Projects
For PhD students: please read the IZW PhD guidelines (\\izw-daten-8\Alle\GUEST\Doktorand(inn)en\IZW-PhD-rules) carefully and follow the instructions (e.g. when you have to give an introduction talk). For any question about these guidelines, contact the PhD coordinators Gábor Czirják (czirjak[at]izw-berlin.de) and Sarah Benhaiem (benhaiem[at]izw-berlin.de).
Meetings
- Regular meetings with your supervisor often help to avoid a waste of time (see also Appendix C). It is in the responsibility of the student to organize and schedule regular meeting with his/ her supervisor or the team.
- You can either arrange a fixed date with your supervisor (e.g. first Monday of every month) or schedule them on demand. Please find an agreement with your supervisor how to handle this best. Please keep in mind that any non-regular meeting should be planned at least two weeks ahead.
- Use the IZW outlook calendar to schedule meetings with supervisors and colleagues (if you have an IZW Email address). That is: create the entry in the calendar and ‘invite’ the others, so that the date/ location appears in each calendar, with sufficient notice time for the automated reminder (i.e. 1 day/ 1 hr).
- Prepare the meetings: Think carefully about your problems/questions and write your agenda (what you want to discuss) before the meeting, make suggestions for your solution(s) and ask the supervisors/collaborators to comment on your solution(s).
- Write the minutes of the meeting and save it in the project folder (we give instructions about this project folder below 3.1.). Minimum information needed: (1) date of meeting, (2) name of participants, (3) questions or problems discussed, (4) main solutions suggested and (5) aims or results to prepare for the next meeting. Write down agreements of the responsibilities of all collaborators.
- Present your project progress regularly in the department meetings, also to get feedback and discuss problems you ran into.
Thesis writing
- Start writing the thesis early enough. Write simple and concise sentences based on what is known in the literature. Join a ‘pub club’ (i.e. retreats for writing), e.g. Team 2 has regular meetings on jointly discussing drafts of each other. Please also read the instructions (Under construction) for how to write good scientific papers.
- Arrange with your supervisor how and in what form you report progress. Getting regular feedback for chapters or even just paragraphs might keep you from running into dead ends. When asking for revisions plan in sufficient time for the reviewers to read and respond (1-3 weeks, depending on the amount of text).
- For PhD students: A first complete manuscript draft should be ready after ~ 1 year after starting time.
- Send the final version of your manuscript/thesis to all supervisors/colleagues at least 4 weeks before the submission date and keep in mind that you might have to revise it.
Organizing Workflows
To store data and results, the IZW follows the DFG-guidelines for good scientific practice. In our department, we often use “scripting” (written code of different programming languages) to process and analyze data, and make results reproducible. Work is structured and organized in projects and each project will get a project ID from the data administrators of the department. We consider a project to be a self-contained topic, e.g. analyses for a manuscript, thesis chapter, etc. For each project a separate folder is created and all relevant scripts, results, the lab-book, documents, literature, minutes of meetings etc. related to this project are to be kept in that main folder. For documents (not for scripts!) use your surname, type of document, project name and in the end the date (YYYYMMDD) as version numbers for file names (kramer_manuscript_lynxibm_20201231.docx) and do not use names like doc_final.docx, doc_finalfinal.docx, doc_lastversion.docx etc. It should contain all information needed to repeat the study and conduct follow-up studies. Whenever possible use the standardized project/folder structure shown in section 3.1. Each project or subproject should have a concise but meaningfully electronic lab-book (see section X.Z) that allows to follow the workflow and the decisions made in it.
Project workflow
Contact the Data Managers (Jan Axtner or Moritz Wenzler-Meya) of the respective teams to organize your project ID, get server access and how to arrange your workspace best. To setup a project folder, we ask you to follow the setup:
- Use the standardized project folder structure! (see Project Folder Structure,
d6R package and GitHub) - Make an outline of the structure separately in your electronic lab-book (see 3.3.1), to indicate what to find in each folder and keep it updated.
- Please hand over the raw data to the data manager before starting your project analysis, to avoid accidental data losses.
- Store the raw data, e.g. data from field work, in the folder named
data-raw. Use a folder namedoutputto store data created from the raw data (e.g. after data cleaning or editing). This should also be the place where themaster tableis located, i.e. the data set of all subsequent analyses. If needed you can have a temporary subfolder in the “output” folder for everyday work and analysis trials. Empty the temporary folder regularly. Plots and figures should be stored in theplotsfolder. Scripts should be stored in theRfolder. Documents like manuscripts, proposals, etc. should be stored in thedocsfolder. Save important, computational or labour intensive interim results or final results. Use appropriate subfolders such asinterim-results_<date>orfinal-results_<date>.
Reproducibility
To ensure reproducibility and for your own sake always try to script your work! If possible use R, Python, SQL, etc. for analyses and avoid manual / mouse commands (e.g. ‘clicking’ in ArcGIS, QGIS etc.). If you cannot use scripts, use tools such as the model-builder in ArcGIS or QGIS and save the drag & drop model builder scripts.
Documentation
Project documentation is not a final task, but an ongoing process starting from day one. DFG guidelines for Safeguarding and Storing of Primary Data states ‘Primary data as the basis for publications shall be securely stored for ten years in a durable form in the institution of their origin.’ Hence, at the end of a research project, you have to hand over a single well-documented folder per project (usually a paper on the respective subject, or the BSc thesis) along with all original data. The project folder should contain all project related documents and data (e.g. simulation model codes, simulation results, scripts for statistical analysis, scripts for tables and figures, manuscripts, important work documents, applications, permits, reports, etc.) as well as a the final version of a thesis or paper. Unfinished work (data from conducted experiments that were not analyzed or published) should contain the above documents as far as possible, including a very detailed method description. For the ease of documentation effort please follow the section (3.3.1).
Electronic lab-book
At the start of each new project, create an electronic lab-book to document your workflow and decisions therein. It is of major importance that you keep this lab-book updated. For the ease of use we recommend simple .doc files (MS Word, Libre Office etc.) to keep record of our work. Write down important thoughts, things you have tried, also failed experiments. Try to use clear and comprehensible notes. Although this generates some additional effort you will realize soon that it will help you and others to track your work and make it reproducible. We highly recommend to use GitHub as version control of your project. You have to connect your GitHub account to the EcoDynIZW organization on GitHub to share code with colleagues. Please create repositories as part of this organization. It is mandatory to hand over the electronic lab-book to the institute as part of the ‘Safeguarding and Storing of Primary Data’ regulation of the German Research Council DFG after finishing your project.
To document scripting we recommend to use R Markdown (.rmd) files if possible. Between your code chunks you should annotate the script with everything that is necessary to understand and follow the code. Additionally, if you push new code to GitHub for version control, you have to comment on your committed code as well. It may be necessary to have an additional word document.
Project Folder Structure
A new project can be started by installing the d6 R-package. Running new_project() with a unique and descriptive name for your project (see below) will create a full scaffolding structure for all your future analysis steps. If you like, you can also specify a path and the project will be created there.
Please create repositories on the EcoDynIZW account and not on your private account!
Root project folder
Name it like:
species|topic_country|simu_method|approach_surname_firstletterofgivenname
e.g. unicornus_wl_sdm_smith_j (unicornus project in wonderland, species distribution model from John Smith) or stability_simu_rpackage_smith_j
- Everything should be written in small letters.
- Avoid spaces or hyphen in the path to the file.
- Species name should be in latin and please use your surname and the first letter of your given name at the end.
- Country should be the international abbreviation.
The root project folder should hold:
- Rproj file: your main file for your R project (if used) — do not confuse this with R-scripts!
- We highly recommend to use R, but if it is necessary to use another programming language and/or program please use the same structure as described in the following:
- data: A folder that contains the complete data, with sub folders for raw (e.g. telemetry data, experiment results, electrophoresis images, vegetation survey plots, species lists, etc.) and processed data. If spatial data is used, placde them in a dedicated geo folder.
- Do not use MS Excel files use .csv or .txt files to store/ save data.
- Metadata belonging to the raw data should be provided in a separate file but it should be given the same name with an additional suffix as the raw data file, e.g. area1_specieslist.txt or area1_specieslist_metadata.txt
- docs: A folder that contains anything related with project administration e.g. applications, permits, grants, timeline, research proposal, the electronic lab-book.
- results: A folder that contains results from your data wrangling and analyses.
- plots: A folder that contains all produced images for the data exploration, presentations, and publications.
- scripts: A folder that contains all scripts that are used within the project.
- If you need additional subfolders add them. Please use also second-level subfolders inside the subfolders to keep a clear and tidy structure.
The main folder structure created in the root directory should look like this:
.
└── unicornus_wl_sdm_smith_j
├── data
├── docs
├── results
├── plots
└── scriptsThe full scaffolding structure including all subdirectories and additional files looks like this:
.
└── unicornus_wl_sdm_smith_j
├── .Rproj.user — Rproject files
├── data — main folder data
│ ├── processed — processed tabular data files
│ │ └── geo — processed geospatial data files
│ └── raw — raw tabular data files
│ │ └── geo — raw geospatial data files
├── docs — documents main folder
│ ├── admin — administrative docs, e.g. permits
│ ├── literature — literature used for parametrization + manuscript
│ ├── manuscript — manuscript drafts (main + supplement)
│ ├── presentations — talks and poster presentations
│ └── reports — rendered reports
├── results — explorative plots, tables etc. (except final figures)
├── plots — final figures for manuscript and supplementary material
├── scripts — script files (e.g. .R, .Rmd, .Qmd, .py, .nlogo)
│ └── zz_submit.R — final script to run before submission
├── .gitignore — contains which files to ignore for version control
├── README.md — contains project details and package dependencies
└── project.Rproj — Rproject file: use to start your projectData Backup
Important: backup your work! Make copies of raw data/files and scripts and save it in another location if you work on private computers. PhD students should work on the M-drive or the U-drive, never directly on the C-drive of the computer.
Manuscript Submissions/Revisions
When you are submitting a manuscript create a copy of the related subproject folder and add a suffix submission_1_<JournalAbbreviation>. This copy shall be stored and never changed—we recommend to zip this folder! When conducting a revisions or resubmission, copy the folder to submission_2_<JournalAbbreviation> and revise the paper.
Please find additional information on ‘how to write a scientific paper’, how to avoid statistical pitfalls or how to organize workflows here.
GitHub repository
The final step after your paper gets published is to make the GitHub repository public (or create a repository if you haven’t before). The repository should contain clean code and data that runs properly. If you are using R, please make sure to use a R-Project with the given folder structure (see appendix A).
The title of the repository should contain the last name of the first author, the year, and the abbreviation of the journal (e.g. Smith_2023_Science). Please add the title of the paper as the description (in quotation marks). Please provide the reference of the paper, including the DOI as a hyperlink, and the abstract in the README file. Add further notes on the files contained and the scripts if needed. Please use this template:
# Smith et al. (2023) *Science*
> **Richard Smith**, Don Joe Radchuk & **Jane Head** (2023): Study title.
*Science* 1-2. DOI: [doi-goes-here](https://doi.org/doi-goes-here)
Abstract text.
## Scripts
... more notes if needed
Add the title in quotation marks as the description and the DOI as link to the repository about (right upper corner). For examples see study repositories on our GitHub).
Finally, make sure to turn the visibility to public and transfer the ownership to the EcoDynIZW GitHub organization. If you have need help or have questions please ask the data manager for help.
Appendices
Appendix A) Scripting
General recommendations
- Write the author, name and date in a header (you can use the d6 package templates for this as well).
- Keep code in scripts as concise as possible.
- Use in-line comments to explain your code that others can follow your code.
- If possible, use dynamic paths (i.e. see R-package
here). - Always set variables/ parameters at the top of your script; do not set values somewhere inside the code as this is error prone, because these values tend to be forgotten to get changed when updating a script. The same applies to loading packages. Please also make sure to only load packages that are really needed and remove those that you are not using for the final analysis.
- Save the output of a script as .rds file to smoothly load it in the next script. Additionally, save the output in a file format usable beyond R, e.g. use .csv or .txt if you want to save tabular data or GeoTiff if you want to save a raster map.
- Use separate scripts to create figures of publishable quality (700 dpi) from the results. Use a vector graphics format like pdf.
- For simulation studies: Include all scripts, e.g. additional model code, batch files with simulated parameters, simulated landscapes and simulation results, and files used for the analyses.
R
- Stick to the Rstudio style guide to ensure readability of code.
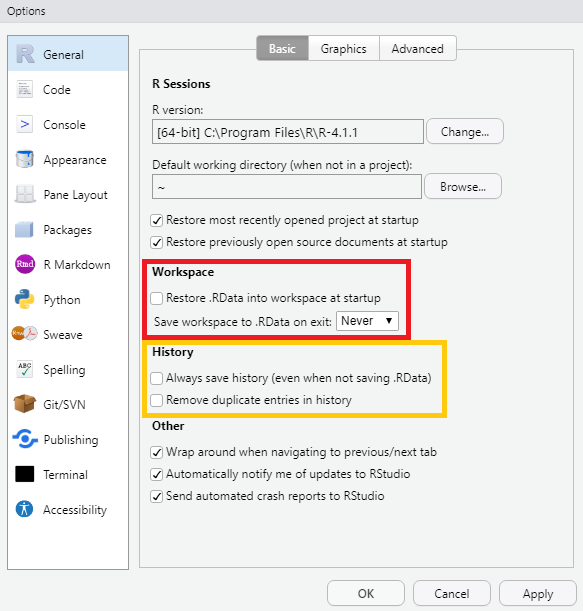
- If you are using RStudio, make sure to set the following under
Tools > Global Options > General > Basic:- untick “Restore .RData into workspace at startup”
- set “Save workspace to .RData on exit:” to “Never”
- untick “Always save history (even when not saving .RData)” (recommended)
- We recommend using R-Studio projects and R Markdown documents when scripting in R. If you use another “integrated development environment” (IDE) make sure you can provide versioned plain R scripts using a relative path to the used data files.
- Implement version control by using GitHub. If you install packages or use scripts and code chunks from GitHub provided by others, be aware to acknowledge the code developer prior to publication of scripts.
- Do not save the R history or the R environment. Always start a fresh and an empty R session to avoid artifacts by using objects of functions that were not defined in your script. More details here: https://www.tidyverse.org/blog/2017/12/workflow-vs-script/.
- If possible use log files to document messages and errors.
Python
In general we recommend using Jupyter notebooks. [under construction]
Netlogo
[under construction]
QGIS
In general we recommend doing GIS analyses in R. If you have to use QGIS use Python as scripting language. However, the RQGIS package provides a great opportunity to script complete workflows in QGIS by using R. Moreover, you can include R scripts in the QGIS toolbox.
[under construction]
Appendix B) Spatial Data
If you are working with spatial data please have the following in mind:
- Always check the projection (cartographic reference system (CRS)) of your data. Depending on what you want to do it could be necessary to change the CRS system, i.e. from an angular unit (like GPS data) to a projected unit in meters. BUT sometimes you cannot easily change it or it will drastically change your data.
- If you want to save you spatial data, please save it as Geopackage (
.gpkg) or raster data GeoTiff (.tif). Please give a clear name, document the filename in your lab book and add the EPSG code of your CRS to the file name. Always useutf-8as file encoding. - If you are not sure how to handle your spatial data please contact Moritz Wenzler-Meya, the GIS manager, for help.
- When using spatial data give clear names for maps ≤ 13 characters without special characters or spaces.
- Write down the cartographic reference system (CRS) at the end of the file name, e.g.
filename_4326. - Always use
utf-8encoding, NEVER use the system setting. If you have to use different character encoding settings, then name the used encoding in the filename. - Save the data as geopackage (
.gpkg, recommended but cannot be used via ArcGis) or shapefile. Geopackages should be the first choice as the format is much quicker in every task (loading, saving, changing and computing), less limited and OGC-Standard (Open Geospatial Consortium). Be aware, that shapefiles have many limitations e.g. max 7 characters for column names. Further information is listed in Appendix B. - Crosscheck your scripts and results. If you have calculated data/processed maps, make a plot and check for consistency. E.g. there should be no data outliers, terrestrial species should not occur in the sea etc. If results are valid, mark it as a milestone in your project documentation.
- Be aware: if you want to change the coordinate reference system (CRS) you have to transform / project the data. Never just specify a new one, because this doesn’t change the CRS!
- Always include the Coordinate Reference System as EPSG code (e.g. 4326 for log/lat WGS84 Coordinates) at the end of a file name, e.g.
my_env_variable_4326.gpkg. - Please save all your geo data into geopackage files (
.gpkg) format to save and process your data. In general, if you export data from R please save it as an R Studio file (.rds). It will be saved without any losses. - Store all point/line/polygon data as WGS84 (EPSG 4326).
- If you do not understand this paragraph, contact the geodata-manager (Moritz Wenzler-Meya) before doing anything involving geodata.
Appendix C) What Nature Says…
Nature 561, 277 (2018)
Why you need an agenda for meetings with your principal investigator
A list of talking points can help with navigating potentially difficult topics and sticky negotiations. As PhD students, we often find ourselves discussing our interactions with our principal investigators (PIs) and swapping advice for improving our mentoring meetings. We have found three practices to be consistently helpful:
- asking our PIs about all aspects of their job;
- preparing an agenda for each meeting;
- negotiating new experiments without explicitly saying ‘no’.
Read the full text: https://doi.org/10.1038/d41586-018-06619-3